-
Notifications
You must be signed in to change notification settings - Fork 43
Skip brain extraction #116
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'll let you keep working on this, but it looks it's taking a good direction.
My main concern is: if the image is skull-stripped, then it is likely to have been INU-corrected too. That makes me think about three issues:
- How would you account for INU-corrected images? I mean, it doesn't hurt to run INU-correction twice (in principle) and let N4 find nothing if the image is intensity corrected.
- The current antsBrainExtraction workflow is modified to recalculate the B1 field using the Atropos segmentation and passing in the white-matter class. Could we also implement something like that here?
- If down the line we decide to implement some B1 field correction in fMRIPrep (and we will certainly do in dMRIPrep), could we break the workflow at construction if some flag (i.e.,
disallow_premasked) is present set true? This way, dMRIPrep could set it on and prevent this behavior to happen while making noise enough to get the attention of the user.
|
Thanks for taking a look.
Any suggestions re: the brain-mask detection though? Or are you happy to leave it as an explicit CLI option? |
cef24eb to
0b90071
Compare
|
I successfully ran ds001110/SherlockMerlin with brain extraction turned off, and the results look decent as far as I can tell. This is ready for another review whenever you get a chance. No rush from me as preprocessing that dataset was my main goal. Here are the reports for the dataset if anyone cares to take a look: https://web.corral.tacc.utexas.edu/neuroscout-datasets/SherlockMerlin/fmriprep/ Main points left to discuss:
cc: @effigies |
|
One more thing: I noticed |
|
Cool. Will have a look now. Should be able to fix the tests by merging/rebasing master. |
|
Hello @adelavega! Thanks for updating this PR. We checked the lines you've touched for PEP 8 issues, and found: There are currently no PEP 8 issues detected in this Pull Request. Cheers! 🍻 Comment last updated at 2020-02-07 20:47:11 UTC |
|
Haven't configured the PEP8 speaks... Ignore the line lengths. |
|
Also, something's come up, and I might not finish a review this afternoon. |
|
update: I didnt realize I think this led to a series of errors which prevented confounds from being extracted. I set it to 10, and that looks pretty close to what you get by threshold a single T1w with 0. A treshold of 1 should also be acceptable. I have no clue what the range of values of T1w images tend to be. I'll re-run with these values. |
83ba99c to
7ee3c6a
Compare
|
I undid the merge master (at least temporarily), because it seems that |
Co-Authored-By: Oscar Esteban <[email protected]>
|
What if we ask users to provide a brain mask? Then we just use it. |
|
But then it would have to be per subject, and I think the brain mask extraction works pretty well (so why create more work for the user?) Given this is fairly niche (although IMO important, given the # of openneuro datasets like this), I think a CLI flag might be a good and easy solution (especially since we would need an over-ride CLI flag anyways, so were adding yet another option no matter what). |
|
Before we all forget about this PR, let me summarize:
So we need to decide if s/fmriprep should either:
Given that the detection could fail either way (although honestly, I think it's probably unlikely), I would vote for 3). I'm also fine with 4), as I think users with pre-stripped datasets may already know it (although I didn't at first, and I was puzzled as to why my results looked bad). |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Alas, I forgot about this PR. But thanks for the summary.
I'd be okay with a --skip-skull-strip flag (or maybe --pre-skull-stripped?) to override detection.
Some additional comments, but otherwise this is looking good. We're going to have merge conflicts with #159. IDK if you want to preemptively merge/rebase on that, or try to squeeze this in and make @mgxd do the work.
Ahem, the latter if possible. |
Co-Authored-By: Chris Markiewicz <[email protected]>
| @@ -329,7 +329,7 @@ def init_single_subject_wf( | |||
| hires=hires, | |||
| longitudinal=longitudinal, | |||
| name="anat_preproc_wf", | |||
| num_t1w=len(subject_data['t1w']), | |||
| t1w=subject_data['t1w'], | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
You need to pass skip_brain_extraction here, and up the chain to the CLI.
|
Need to update the niworkflows dependency here: |
|
Pulling from a resolved comment:
|
smriprep/cli/run.py
Outdated
| @@ -95,6 +95,9 @@ def get_parser(): | |||
| g_ants.add_argument('--skull-strip-fixed-seed', action='store_true', | |||
| help='do not use a random seed for skull-stripping - will ensure ' | |||
| 'run-to-run replicability when used with --omp-nthreads 1') | |||
| g_ants.add_argument( | |||
| '--skip-brain-extraction', required=False, action='store_true', | |||
| default=False, help='Skip the brain extraction workflow (for pre-extracted T1ws))') | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hmm. This takes care of false negatives, but I suppose we should consider false positives, where we might incorrectly decide an image is skull-stripped.
Maybe we want --skull-strip {auto,skip,force}?
Or am I over-thinking the problem?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yeah, I previously suggested this, but also worried I was overthinking it. False positives are actually probably a bigger problem in a way (many more people have normal T1s)
If you're comfortable with the additional CLI complexity, I think it this makes the most sense.
Again, we could also remove the detection if we think its not reliable, and only have a skip detection flag.
My preferences in order: 1) CLI argument w/ 3 options 2) skip only CLI flag, with no detection 3) skip flag + detection (current)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I lean toward 1 as well.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@mgxd ? any opinion
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
i'm fine with the 3 way approach, with the default being auto. It should definitely document which route was taken
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Sure, how? You mean which option was select or what auto detected? And if so, where do I log that?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
cool! couple comments
smriprep/workflows/anatomical.py
Outdated
| @@ -22,15 +25,17 @@ | |||
| ) | |||
| from niworkflows.interfaces.images import TemplateDimensions, Conform, ValidateImage | |||
| from niworkflows.utils.misc import fix_multi_T1w_source_name, add_suffix | |||
| from niworkflows.anat.ants import init_brain_extraction_wf | |||
| from niworkflows.anat.ants import ( | |||
| init_brain_extraction_wf, init_n4_only_wf, _pop | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
not a huge fan of importing "private" functions
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Sure, it's just a pop function so we can redefine.
| templateflow |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
why is this different? spacing?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
not sure 🤷
smriprep/cli/run.py
Outdated
| @@ -95,6 +95,9 @@ def get_parser(): | |||
| g_ants.add_argument('--skull-strip-fixed-seed', action='store_true', | |||
| help='do not use a random seed for skull-stripping - will ensure ' | |||
| 'run-to-run replicability when used with --omp-nthreads 1') | |||
| g_ants.add_argument( | |||
| '--skip-brain-extraction', required=False, action='store_true', | |||
| default=False, help='Skip the brain extraction workflow (for pre-extracted T1ws))') | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
i'm fine with the 3 way approach, with the default being auto. It should definitely document which route was taken
|
Looks like @mgxd got in first. Merge/rebase? |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm gonna cut a new release in order to get an fmriprep release out, but this is close. I can clean up the git history once all the above is addressed 👍
Co-Authored-By: Mathias Goncalves <[email protected]>
…into enh/skull_strip_skip
|
Closed in favor of #167. |
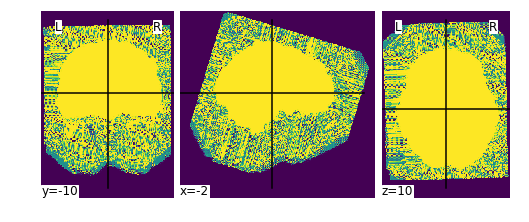
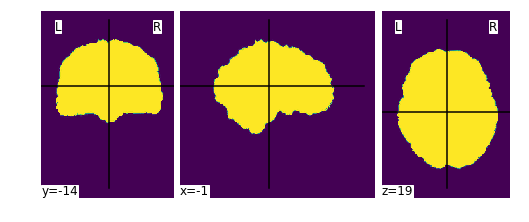
Fixes #12 by creating an alternative workflow to
init_brain_extraction_wf,init_n4_only_wf.This wf only does n4 bias field correction, and creates a mask file by thresholding the input T1w image.
My main concern is if this is the right approach.
Alternatively, the
init_brain_extraction_wfworkflow could be altered in niwokflows, but as this is supposed to be an implementation of ANTS' brain extraction, I thought it was better not to touch that.I also though about just changing the
init_anat_preproc_wffunction to change the connections based on if brain extraction would be run, but that got messy quick.Also, I'm currently just using a parameter to indicate if brain extraction should be skipped. Although the function to detect if brain extraction is needed is easy, I'm not sure how to dynamically create a workflow based on the output of a Node.
Any pointers here?